About
Our lab aims at understanding how genetic changes between individuals can or cannot result in disease by quantifying the impact mutations have on protein aggregation and toxicity.
We are particularly interested in amino acid sequences that can adopt different conformations and undergo a process of self-assembly which results in distinct physical states.
The aggregation of proteins into insoluble amyloid fibrils is a key process in the pathogenesis of a number of neurodegenerative conditions, such as Parkinson’s disease or Amyotrophic Lateral Sclerosis. However, examples of functional amyloid are also widespread in nature, especially across bacteria and fungi. Our work aims at systematically deciphering the sequence-dependencies of the process of aggregation in both functional and pathological contexts.
Recently, it has become clear that proteins can also self-assemble into a more dynamic and reversible state through a process of liquid de-mixing which is thought to contribute to the organization of the intracellular space. However, also for proteins undergoing liquid de-mixing, the balance between function and dysfunction is far from clear. It is also unknown if, in vivo, liquid de-mixed states are precursors of insoluble amyloid-like states, and to which extent proteins are structured once in the liquid state.
How we do it
In order to understand how mutations affect these delicate equilibria and to elucidate when and why a sequence starts aggregating or becomes toxic for the cell, our lab integrates experimental and computational approaches in different model systems. Recently, we have developed massively parallel approaches based on Deep Mutational Scanning (DMS) to quantify the toxicity or the aggregation propensity of hundreds of thousands of protein sequences in vivo. We believe that by portraying the full landscape of the effects of mutations in a specific protein domain we can reach a more systematic and comprehensive understanding of the determinants of amyloid formation and toxicity.
We are also interested in developing similar high-throughput strategies to measure in vivo the effect of mutations on the physical state the proteins acquire upon mutation (diffuse, liquid de-mixed, insoluble) and to study the interactions between mutations to report on the conformations proteins adopt as they self-assemble. Overall, the exhaustive datasets we are generating will give mechanistic insights on the process of protein aggregation, while also reporting on specific conformations and mechanisms leading to cellular toxicity. We also aim at using the datasets we generate to develop novel predictors of protein aggregation.
We focus on all classical amyloids, such as the amyloid-beta peptide, the main component of the plaques found in Alzheimer’s disease patients, but also on functional amyloids and on a less characterized part of the human proteome which is able to undergo liquid de-mixing: prion-like domains. Just like all disordered protein regions, prion-like domains are particularly difficult to study in vitro. In this perspective, in vivo approaches such as the ones we develop, can provide a unique opportunity to investigate these sequences in a systematic way.
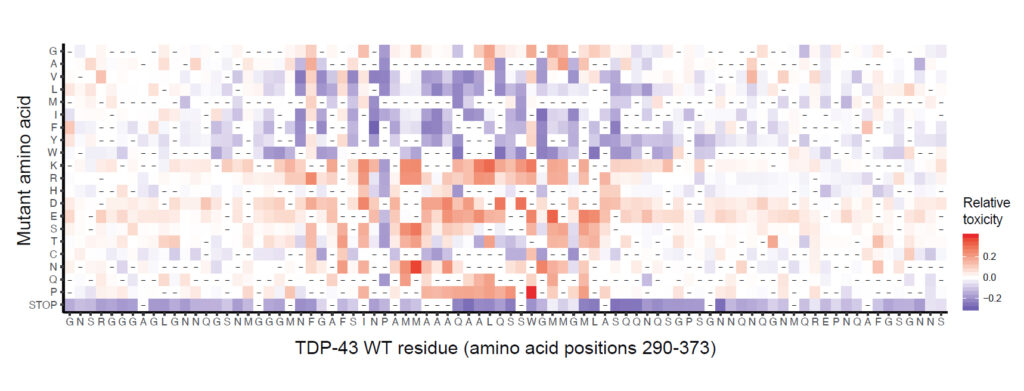
Map of the effect of mutations on toxicity of the TDP-43 Prion-like Domain.
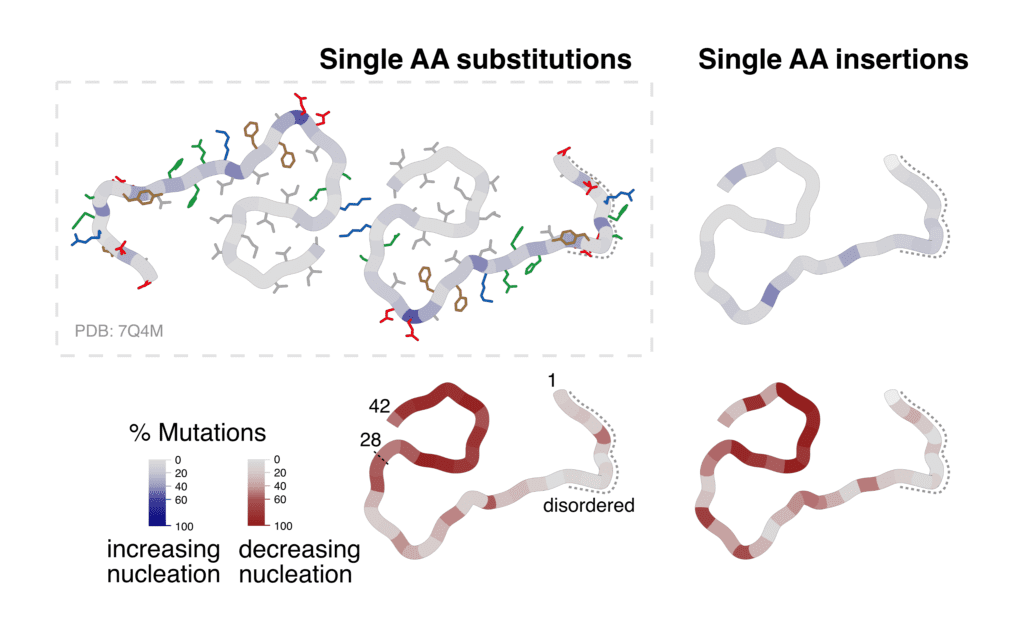
Percentage of substitutions and insertions increasing or decreasing amyloid formation of the Amyloid-Beta peptide, visualized on the cross-section of ex-vivo fibrils (7Q4M).
Staff
Projects
| NATIONAL GRANTS | FINANCER | PI |
|---|---|---|
| AMYNDEL · Deciphering the consequences of different types of genetic variation in amyloid forming sequences by deep mutagenesis ( 2022-2025) | MICIU · Generación Conocimiento: Proyectos I+D | Benedetta Bolognesi |
| DeepAmyloids · Massively parallel mutagenesis to understand, predict and prevent amyloid nucleation in neurodegenerative diseases (2021-2024) | Obra Social La Caixa | Benedetta Bolognesi |
| FINISHED PROJECTS | FINANCER | PI |
|---|---|---|
| Poly-STOP · Developing modulators of protein aggregation in polyglutamine diseases by deep mutational scanning (2021-2022) | BIST · Barcelona Institute of Science and Technology | Benedetta Bolognesi |
| PRIOMUT · Escaneado exhaustivo de mutaciones en un dominio priónico para entender la toxicidad inducida por proteínas (2019-2021) | MICIU / Retos investigación: Proyectos I+D | Benedetta Bolognesi |
Publications
Equipment
- Thermo MaxQ 8000
Collaborations
- Priyanka Narayan
NIH-NIDDK - Xavier Salvatella
IRB, Barcelona - Fran Supek
IRB, Barcelona - Ben Lehner
CRG, Barcelona - Luke McAlary /Justin Yerbury
University of Wollongong, Australia
News
BIST Forum, a meeting to highlight the value of frontier research
The BIST Forum has discussed how excellent science enhances the development of society and economic growth. The event was attended by the President of the Catalan Government, the Mayor of Barcelona, the heads of the highest economic institutions and the rectors of the main universities. At the event, the new BIST IGNITE projects for multidisciplinary research were announced, three of which have the participation of IBEC.
Benedetta Bolognesi awarded a prestigious European ERC Consolidator Grant
The researcher at the Institute for Bioengineering of Catalonia has been awarded an ERC Consolidator Grant. This prestigious European funding supports excellent scientists and scholars who are consolidating their independent research teams to pursue their most promising scientific ideas. The €2 million grant over 5 years will allow Bolognesi and her team to develop a new method for identifying mutations that lead to the formation of amyloids—aggregates of proteins that contribute to a variety of diseases, including Alzheimer’s and Parkinson’s.
A new atlas unveils which mutations are behind the noxious amyloid formation
Researchers from the Institute for Bioengineering of Catalonia (IBEC) have developed the most complete atlas up to date of the genetic mutations that cause the formation of amyloid beta fibril, … Read more
Science and Art: IBEC collaborates on a work by Antoni Muntadas at Ars Electronica 2022
When is a living organism considered extinct? This is one of the fundamental questions of the artwork presented at Ars Electronica 2022 by Antoni Muntadas, a Catalan artist based in … Read more
Postdoctoral researcher at the Protein Phase Transitions in Health and Disease Research group
Ref: Postdoctoral / Deadline: 24th August
Lab Technician at the Protein phase transitions in health and disease Research Group
Ref: Lab Technician / Deadline: 29th July 2022
Benedetta Bolognesi and Ben Lehner win competitive funding to join forces against neurodegenerative diseases
The “la Caixa” Foundation will fund a large and innovative research project co-led by IBEC Junior Group Leader Benedetta Bolognesi and by ICREA research professor Ben Lehner at CRG, which aims to gain a better understanding of the genetic causes leading to neurodegenerative diseases. Researchers will combine deep mutagenesis and machine learning techniques to produce a “map of dementia” as a method to predict whether a person is more susceptible to suffer these diseases.
IBEC researchers win two “BIST Ignite Seed Grants”
IBEC researchers receive two “Ignite Seed Grants” to combine their skills with other BIST members to seek scientific answers to health challenges. Benedetta Bolognesi will study, with the IRB, Huntington’s disease and other neurodegenerative pathologies without treatment. On the other hand, Juan Manuel Fernández-Costa and researchers at ICFO will develop muscles-on-a-chip and biomagnetism sensors to accelerate the design of new treatments for muscular dystrophy.
One more step towards early detection of Alzheimer’s
Benedetta Bolognesi, junior group leader at IBEC, appears in different media for a recent study published in the eLife magazine. In the study they show the first map with all the possible mutations in the amyloid beta peptide and tested how they influence its aggregation into plaques, a pathological hallmark of Alzheimer’s disease.
First comprehensive map of amyloid plaque mutations opens new avenues for early detection of Alzheimer’s disease
A study published in the journal eLife made all the possible mutations in the amyloid beta peptide and tested how they influence its aggregation into plaques, a pathological hallmark of Alzheimer’s disease.
Jobs
Research Assistant at the Protein Phase Transitions in Health and Disease Research Group (RA_BB)
Ref: RA_BB // Deadline: 26/01/2024
Senior Laboratory Technician at the Protein Phase Transitions in Health and Disease Research Group (SLT_BB)
Ref: SLT_BB // Deadline: 11/01/2024
Research Assistant at the Protein Phase Transitions in Health and Disease Research Group
Ref: RA_BB // Deadline: 04/08/2023
Research Assistant at the Protein Phase Transitions in Health and Disease Research Group
Ref : RA_BB // Deadline 28/04/2023
Postdoctoral researcher at the Protein Phase Transitions in Health and Disease Research group
Ref: Postdoctoral / Deadline: 24th August
Lab Technician at the Protein phase transitions in health and disease Research Group
Ref: Lab Technician / Deadline: 29th July 2022
2 Postdoctoral researchers at the Protein Phase Transitions in Health and Disease Research Group
Application Deadline: 24/10/2021Ref: PD-BB The Protein phase Transitions in Health and Disease group at the Institute for Bioengineering of Catalonia (IBEC) is looking for two Postdoctoral Researchers to develop deep mutagenesis projects in the context of amyloid forming proteins. The contract will be within the framework of a larger project funded by La Caixa.


 ibecbarcelona.eu
ibecbarcelona.eu